Multiscale learning framework revolutionizes structural biology and accelerates drug discovery
Meta Information
Meta Title: AI Protein Mapping Breakthrough 2025: 50% Faster Predictions
Meta Description: Scientists develop revolutionary multiscale AI system for protein structure prediction. New method achieves unprecedented accuracy while cutting computational costs. Major breakthrough for drug discovery.
Focus Keywords: protein structure prediction, AI protein mapping, multiscale learning, drug discovery, computational biology, AlphaFold, structural biology
Table of Contents
- Executive Summary
- The Core Challenge
- The Breakthrough Solution
- How It Works
- Real-World Applications
- The Broader AI Revolution
- Current Limitations
- Future Implications
Executive Summary
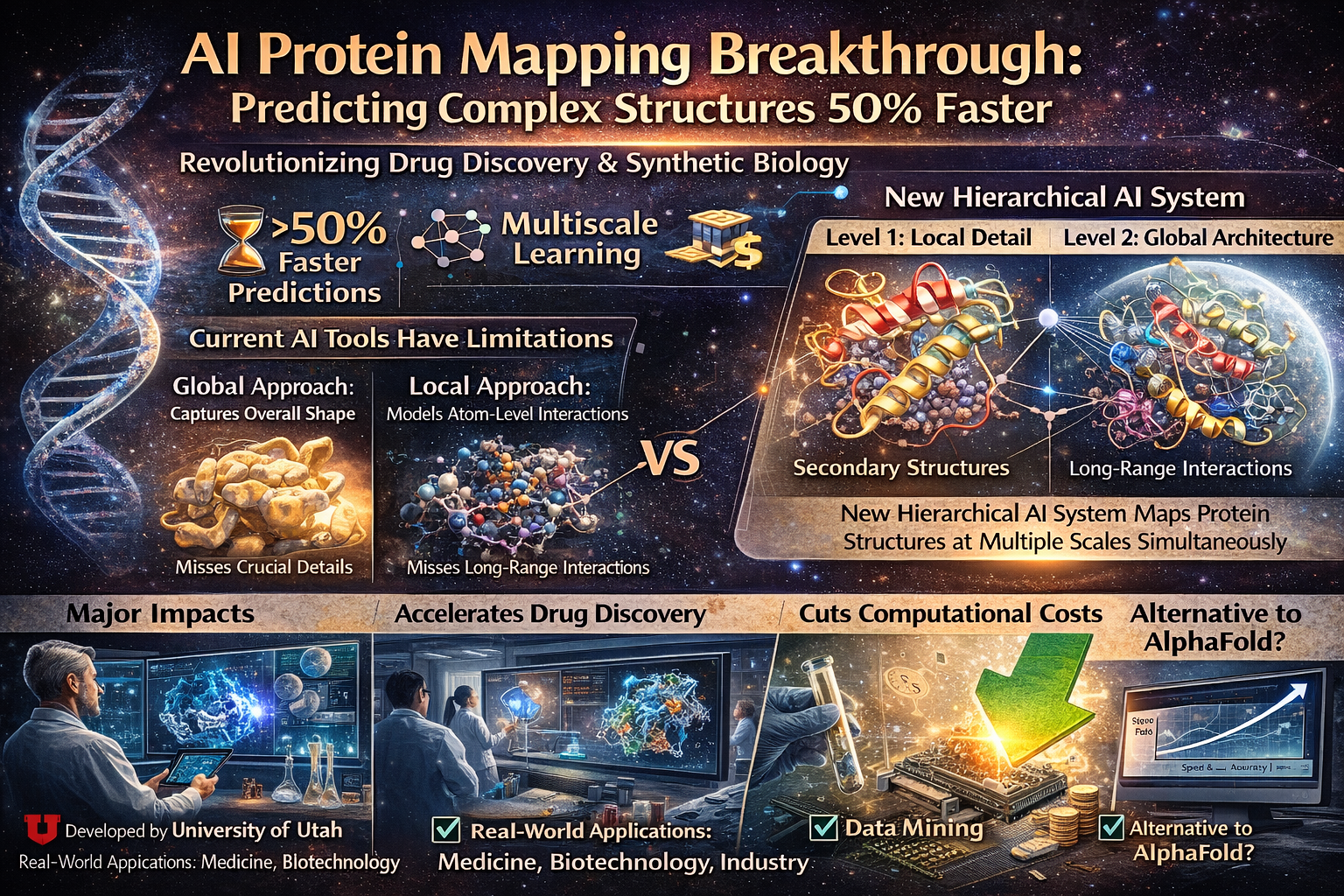
Scientists from the University of Utah’s Department of Mathematics have developed a groundbreaking multiscale graph-based learning framework that transforms how researchers map complex protein structures.
Working alongside colleagues from UCLA and UC Davis, the team addressed fundamental limitations in existing protein modeling methods by creating a hierarchical approach that captures microscopic details and global structural patterns simultaneously.
Key Achievements:
- 50% reduction in computational costs
- Improved prediction accuracy on established benchmarks
- Better handling of complex multi-domain proteins
- Maintains maximum structural information without data loss
- Scales efficiently to larger protein structures
This advancement arrives during an unprecedented era in protein science, where artificial intelligence is revolutionizing our understanding of molecular biology.
The Core Challenge
Why Protein Structure Matters
Proteins are nature’s molecular machines. They perform virtually every function in living cells—from digesting food to fighting infections to repairing DNA damage.
Understanding how proteins fold into their three-dimensional shapes is critical for:
- Developing new medicines
- Treating genetic diseases
- Understanding disease mechanisms
- Creating industrial enzymes
- Designing new biomaterials
But there’s a fundamental problem.
The Trade-Off Problem
Traditional computational approaches face a critical dilemma:
Option 1: High Detail, Limited Scope Methods capturing fine atomic details often miss long-range interactions crucial for protein function. They see the trees but miss the forest.
Option 2: Big Picture, Lost Precision Approaches modeling global structure sacrifice local precision. They see the forest but miss important details about individual trees.
This limitation has real consequences. Consider prion proteins—the agents behind mad cow disease and related conditions. These proteins can fold into dramatically different shapes from identical amino acid sequences. One shape is benign, the other pathogenic. The difference lies entirely in spatial arrangement.
As recent research published in Frontiers in Pharmacology explains, representing proteins at the atomic level incurs significant computational costs, while residue-level representations often fail to capture critical multiscale features like secondary structures that play fundamental roles in protein folding.
The Breakthrough Solution
A Hierarchical Approach
The University of Utah team’s innovation mirrors how biology itself organizes proteins.
Their framework constructs a hierarchical graph combining:
- Detailed subgraphs representing secondary structural motifs (α-helices, β-strands)
- Coarse-grained graphs illustrating spatial arrangement
- Dedicated neural networks for both local and global feature learning
Think of it like mapping a city. You need:
- Street-level detail (which buildings connect to which)
- Bird’s-eye view (how neighborhoods relate across the metro area)
Previous methods forced researchers to choose one or the other. This new approach provides both simultaneously.
Why This Matters
The modular design demonstrably improves prediction accuracy and reduces computational demands across established benchmarks. According to the research team, this represents a significant advance in addressing limitations that have plagued the field for decades.
The researchers proved their hierarchical framework preserves maximal expressiveness—ensuring no critical structural information is lost during processing—while simultaneously reducing computational overhead by approximately 50%.
How It Works
The Two-Tiered Architecture
Layer 1: Local Detail Processing The first neural network operates within individual secondary motifs to capture interactions between amino acids. It examines:
- α-helices (spiral structures)
- β-strands (flat sheets)
- Loops and turns
- Local chemical interactions
Layer 2: Global Architecture Mapping The second neural network models higher-level relationships across these motifs, capturing:
- Long-range interactions
- Domain arrangements
- Overall protein topology
- Spatial relationships between distant regions
Technical Innovation
This dual-network approach solves several problems simultaneously:
- Computational Efficiency: By processing details hierarchically, the system avoids redundant calculations
- Information Preservation: No critical structural data is lost in translation
- Scalability: The method handles large, complex proteins more effectively
- Accuracy: Testing shows improved performance on challenging benchmarks
Real-World Applications
Drug Discovery Acceleration
More accurate protein modeling isn’t just academic achievement. The implications touch virtually every corner of medicine and biotechnology.
Understanding precise protein structures reveals where potential drugs can bind. This knowledge helps pharmaceutical companies:
- Identify therapeutic targets faster
- Design more effective medications
- Predict drug side effects earlier in development
- Reduce development costs and timelines
Current drug development takes 10-15 years and costs billions. Better protein modeling could cut years off this timeline.
According to a comprehensive review in Computers in Biology and Medicine, AI-driven protein structure prediction is already significantly impacting drug discovery, protein engineering, and disease research.
Custom Protein Engineering
Scientists can now design proteins that never existed in nature. Research published in PMC describes how de novo protein design is revolutionizing synthetic biology by enabling first-principle rational engineering unbound by evolutionary constraints.
Recent successes include designed proteins that protect mice entirely from lethal toxin doses, with high-affinity binding designs achieving nanomolar-range dissociation constants.
Potential applications include:
- Biosensors detecting environmental pollutants
- Enzymes breaking down plastic waste
- Therapeutic proteins treating genetic diseases
- Industrial catalysts for green chemistry
- Next-generation vaccines
Disease Understanding
Many diseases stem from protein misfolding or malfunction. Better structural models help researchers:
- Understand disease mechanisms at molecular level
- Identify what goes wrong in genetic disorders
- Design therapies targeting specific protein defects
- Predict which genetic mutations cause problems
For example, Alzheimer’s disease involves the accumulation of misfolded proteins. Understanding these structures precisely could lead to breakthrough treatments.
The Broader AI Revolution
AlphaFold’s Foundation
This breakthrough arrives during an extraordinary period in AI-powered protein science.
AlphaFold2, developed by Google DeepMind, achieved near-experimental accuracy for 98.5% of human proteins. This performance in the CASP14 competition is considered the second-largest breakthrough in life sciences after the human genome project.
AlphaFold 3 expanded capabilities by jointly modeling proteins with DNA, RNA, small molecules, ions, and post-translational modifications—delivering 50% or greater accuracy improvement on protein-ligand and protein-nucleic acid interactions.
Complementary Advances
Other research teams are pushing boundaries too:
D-I-TASSER: This tool combines artificial intelligence with physics-based simulations to predict complex protein structures about 13% more accurately than previous state-of-the-art methods, particularly excelling at challenging multi-domain proteins.
AlphaSync: Researchers at St. Jude Children’s Research Hospital created a continuously updated database maintaining 2.6 million predicted protein structures across hundreds of species. When they first launched, they found a backlog of 60,000 outdated structures.
Protein-Protein Interaction Prediction: According to a recent review in Molecular Therapy, AI-based approaches are transforming our ability to predict how proteins interact—crucial for understanding cellular processes and designing drugs.
Current Limitations
Despite remarkable progress, significant challenges remain.
Dynamic Behavior
Proteins aren’t static sculptures. They’re dynamic machines that change shape as they work.
As noted in the Journal of Chemical Information and Modeling, current AI tools excel at predicting single snapshots but struggle with:
- Multiple conformational states
- Protein folding pathways
- Dynamic transitions
- Flexible regions
Accuracy Gaps
AlphaFold3 still faces limitations with approximately 4.4% of predictions exhibiting structural errors, particularly prominent in large complexes. The focus remains primarily on static conformations rather than capturing protein dynamic transitions.
Generalization Questions
The generalizability of new multiscale frameworks to entirely novel protein structures remains an area for continued investigation. Computational efficiency and scalability with larger datasets warrant further study.
Special Cases
Significant challenges remain in predicting:
- Host-pathogen interactions
- Interactions between intrinsically disordered regions
- Immune response-related interactions
- Membrane proteins
- Multi-domain complexes
Future Implications
Next-Generation Goals
Future AI models might simultaneously predict:
- Protein structure
- Dynamic behavior
- Binding affinity
- Enzymatic activity
- Specificity profiles
The ultimate goal involves modeling entire cellular machines and pathways—not just individual proteins but complete biological systems.
Integration and Convergence
The field is moving toward unified models that combine:
- Structure prediction
- Dynamic simulation
- Functional annotation
- Interaction mapping
- Drug binding prediction
As highlighted in a Royal Society Interface review, the AlphaFold revolution has, if anything, increased the need for chemical and structural literacy among biologists to properly interpret predictions.
Democratization of Research
Lower computational costs mean smaller labs and institutions can participate in cutting-edge research. This democratization accelerates discovery across the entire field.
According to Science Publishing Group research, AI-driven protein structure and function prediction tools are making complex biological data accessible to many research groups, accelerating discovery without expensive and time-consuming experiments.
Why This Breakthrough Matters Now
The Perfect Storm
Several factors converge to make this the ideal moment for such advances:
1. Data Availability The Protein Data Bank (PDB) now contains structures for hundreds of thousands of proteins. Machine learning thrives on large datasets.
2. Computational Power Cloud computing and specialized AI chips make sophisticated calculations accessible to more researchers.
3. Algorithm Innovation New architectures like graph neural networks and transformers unlock capabilities impossible with previous approaches.
4. Interdisciplinary Collaboration Mathematicians, computer scientists, and biologists working together produce better results than any field alone.
Practical Impact Timeline
Near-term (1-3 years):
- Faster drug target identification
- More efficient protein engineering workflows
- Better understanding of disease mechanisms
- Improved experimental design
Medium-term (3-7 years):
- First medications designed using these tools reach clinical trials
- Industrial enzymes with custom properties
- Diagnostic tools based on protein interactions
- Personalized medicine applications
Long-term (7+ years):
- Complete cellular pathway models
- Designer proteins for environmental remediation
- Revolutionary materials based on protein structures
- Synthetic organisms with custom capabilities
The Bigger Picture: Rewriting Biology’s Code
The protein folding problem—once considered one of biology’s grand challenges—is rapidly being solved. This opens entirely new chapters in humanity’s relationship with biology.
Paradigm Shifts
We’re moving from:
- Reading life’s code → Writing new programs
- Understanding natural proteins → Engineering molecular machines
- Treating symptoms → Fixing molecular causes
- Discovering compounds → Designing therapeutics
- Observing systems → Predictively modeling life
Beyond Medicine
The implications extend far beyond healthcare:
Agriculture: Crops with enhanced nutrition or climate resistance Materials Science: Self-assembling structures and smart materials Environmental Protection: Proteins that break down pollutants or capture carbon Manufacturing: Biological factories producing chemicals sustainably Energy: Improved biofuels and biological solar cells
What Researchers Are Saying
The scientific community recognizes this as a transformative moment.
As noted in the RCSB Protein Data Bank documentation, protein structures elucidated through various methods continue to expand our understanding, but AI predictions are now complementing experimental approaches in unprecedented ways.
Leading structural biologists emphasize that while these tools are powerful, they don’t replace experimental validation. Instead, they guide experiments more efficiently and help interpret results more effectively.
Actionable Insights
For Researchers
If you’re working in related fields:
- Explore open-access databases like AlphaFold Database
- Consider how hierarchical modeling might apply to your problems
- Collaborate across disciplines—biology, computer science, mathematics
- Validate computational predictions experimentally when possible
- Stay current with rapid developments in the field
For Industry
Pharmaceutical and biotech companies should:
- Invest in computational infrastructure
- Train staff in AI-assisted drug discovery
- Integrate structure prediction into development pipelines
- Partner with academic institutions on cutting-edge research
- Prepare for dramatically accelerated discovery timelines
For Policy Makers
Support for this field requires:
- Funding for interdisciplinary research programs
- Investment in computational infrastructure
- Education programs combining biology and computer science
- Regulatory frameworks adapted to AI-assisted drug development
- International collaboration on protein structure databases
Conclusion: A Molecular Revolution Accelerates
The University of Utah team’s multiscale graph-based approach exemplifies how clever architectural choices unlock breakthrough capabilities. By respecting biology’s hierarchical organization, they’ve created a tool that’s both more accurate and more efficient.
This isn’t incremental progress. Combined with advances like AlphaFold and D-I-TASSER, these tools fundamentally transform humanity’s ability to understand and manipulate life’s molecular machinery.
The protein folding problem took decades to crack. Now, with AI’s help, scientists are moving beyond prediction to design—creating proteins that never existed, solving problems nature never encountered, and opening therapeutic possibilities previously confined to science fiction.
The Next Chapter
As Nature’s protein structure prediction portal demonstrates, millions of protein structure models have been generated in a remarkably short time. This wealth of new structural information demands careful interpretation and creates unprecedented opportunities.
The molecular revolution isn’t coming. It’s here. And it’s accelerating.
Additional Resources
Primary Research Databases
- Protein Data Bank (RCSB PDB) – Global archive for 3D protein structures
- AlphaFold Database – 200+ million predicted structures
- AlphaSync – Continuously updated predictions
- UniProt – Comprehensive protein sequence database
Key Research Papers
- Frontiers in Pharmacology: Deep Learning Review
- Computers in Biology: AlphaFold Analysis
- Royal Society Interface: AlphaFold Revolution
- Molecular Therapy: Protein-Protein Interactions
Educational Resources
- Nature Reviews: Structural Biology
- NIH: Advances in AI for Cancer Drug Discovery
- Science Publishing Group: AI in Protein Science
Tools and Platforms
- AlphaFold – Google DeepMind’s prediction tool
- AlphaFold 3 – Latest version
- D-I-TASSER – Structure prediction server